Carpet beetle enzymes for sustainable valorisation of keratin
- Project lead
- Federico Sabbadin
- Institute
- University of York
Summary:
Keratin is an abundant structural protein that forms the bulk of hair, fur, feathers, hooves, horns and scales in animals. Over 65 million tons of keratin waste are generated each year from the food and textile industry, and they represent a cheap and plentiful source of building blocks (peptides and amino acids) for the production of fertilisers, food supplements, biogas, biodegradable food packaging, composite materials and textiles.
Keratins are stabilised by abundant inter and intra-chain disulphide bonds, as well as hydrogen, hydrophobic and ionic bonds that confer high resistance to acids, alkaline reagents, organic solvents and conventional proteases. Due to its high recalcitrance to degradation, most keratin is simply buried in landfills or incinerated, leading to land pollution and greenhouse gas emissions. Keratin waste thus represents a major hazard to both the environment and human health.
Until now, keratin deconstruction has been achieved mainly through harsh physico-chemical treatments (requiring high temperature and pressure, toxic chemicals and solvents) that are not environmentally friendly and are not compatible with a circular, sustainable bioeconomy. Here we propose to use a multidisciplinary approach to identify keratin-degrading enzymes produced by carpet beetles, which have naturally evolved to digest keratin. This project will produce carpet beetle keratinases in pure form, test their ability to deconstruct wool and feathers, and pave the way to the creation of efficient keratin-degrading enzyme cocktails for industry.
Aims:
The project had four key objectives:
- Isolate high-quality RNA and digestive proteins from the carpet beetle Anthrenus flavipes fed on wool;
- 2- Build a de novo transcriptome of A. falvipes and carry out shotgun proteomics of its digestive fluids to identify key enzymes involved in keratin digestion;
- 3- Produce recombinant A. flavipes enzymes in yeast;
- 4- Biochemically characterise the purified enzymes and test their ability to deconstruct keratin into peptides and amino acids for downstream applications.
Outcomes:
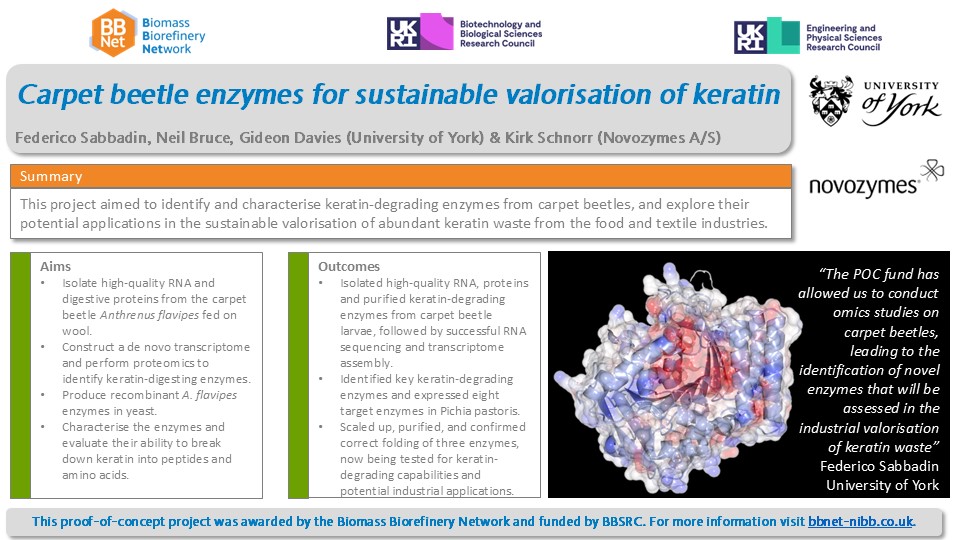
We have achieved several milestones in our study of carpet beetle larvae fed on wool and oats. First, we successfully isolated high-quality RNA and protein from the larvae’s digestive systems and purified larval digestive fluids containing keratin-degrading enzymes. Next, we conducted RNA sequencing, assembled a novo transcriptome, and identified the most expressed and induced carpet beetle genes during keratin feeding. Shotgun proteomics of the beetle`s digestive fluids further revealed the most abundant and enriched enzymes. Our findings highlighted enzymes capable of breaking disulfide bonds. hydrolysing the keratin backbone, and degrading embedded lipids and waxes. Additionally, we identified induced enzymes which may play roles in attacking specific post-translational modifications of keratin. Finally, we cloned and expressed the coding sequences in the yeast Pichia pastoris. This process yielded soluble expression of eight enzyme targets, including thiol reductases, proteases, and amidases. Three enzymes were successfully scaled up, purified, and confirmed for correct folding through a thermal shift assay. We are now exploring their keratin-degrading capabilities and assessing their potential applications in the industrial conversion of keratin waste into high-value peptides and amino acids.
Impact:
This project advanced our understanding of keratin digestion in carpet beetles, generating high-quality omics data and identifying key enzymes. We demonstrated that Pichia pastoris is an effective heterologous expression system for these sequences, and purified three folded targets to high yield, paving the way for converting keratin waste into valuable peptides and amino acids.
Academic partners: Federico Sabbadin, University of York, Neil Bruce, University of York, Gideon Davies, University of York
Industrial partners: Kirk Schnorr, Novozymes A/S

Figure 1. Larvae of carpet beetle (Anthrenus flavipes) reared at CNAP.




